In the example BLAST below, there is only one real hit which is the first one.
You should see a match to the entire flanking sequence tag, except for a few bases which may be due to sequencing errors.
We use a default e-value of 1e-20 on our blast page, which would normally only return a result like the first one.
In this example, we used a less stringent e-value of 1e-04 to demonstrate the range of results that can be returned by the blast program.
If you see a match to only a small portion of the sequence, then it is not likely that there is a insertional mutant matching your gene of interest.
![]()
BLASTN 2.2.9 [May-01-2004]
Reference:
Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schäffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Database: combined_genbank_submitted
10,808 sequences; 4,449,403 total letters
Query= (1296 letters)
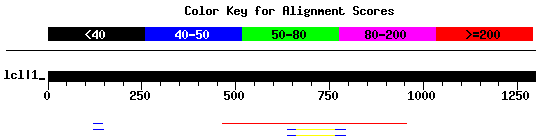
Score E Sequences producing significant alignments: (bits) Value N RGT4508_5.1 973 0.0 1 RGT4569_5.1 44 1e-05 2 RGT4568_5.1 44 1e-05 2 RdSpm2990B_3.1 48 4e-05 1 RGT3379_5.1 46 1e-04 1>RGT4508_5.1 Length = 491 Score = 973 bits (491), Expect = 0.0 Identities = 491/491 (100%) Strand = Plus / Plus Query: 465 ggtcgcgctgcgctgcgaggatccgtgtgagggttactgcttcgagtgtgaggattcatt 524 |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| Sbjct: 1 ggtcgcgctgcgctgcgaggatccgtgtgagggttactgcttcgagtgtgaggattcatt 60 Query: 525 ggcaattgagagccagatggtggccgatgatggcgccggaggcggcgaggaagggtatgg 584 |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| Sbjct: 61 ggcaattgagagccagatggtggccgatgatggcgccggaggcggcgaggaagggtatgg 120 Query: 585 atgtgttgtcacagggatgccgaatcttgggaacacctgctacctgaacgcgctgctgca 644 |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| Sbjct: 121 atgtgttgtcacagggatgccgaatcttgggaacacctgctacctgaacgcgctgctgca 180 Query: 645 gtgcctccttgtgctcgggaagctgcgggcgaggatcttggggccgggcgctccgtcggg 704 |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| Sbjct: 181 gtgcctccttgtgctcgggaagctgcgggcgaggatcttggggccgggcgctccgtcggg 240 Query: 705 tgtccttggtgacttgctgcatgatctctttgttggtacaaatggtccgagctacgctcg 764 |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| Sbjct: 241 tgtccttggtgacttgctgcatgatctctttgttggtacaaatggtccgagctacgctcg 300 Query: 765 acgcctgctggacccggcgatgctcttgcgttgtgtgcgctttcgttacccgcagttccg 824 |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| Sbjct: 301 acgcctgctggacccggcgatgctcttgcgttgtgtgcgctttcgttacccgcagttccg 360 Query: 825 aggcattgctatgcaggactgtcatgaattgctttgctgtttgcgcgatggtttggatga 884 |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| Sbjct: 361 aggcattgctatgcaggactgtcatgaattgctttgctgtttgcgcgatggtttggatga 420 Query: 885 ggatgaaaggaaatggagggctggtaagatgcaacaaggtgctcctagtgctgtggctcc 944 |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| Sbjct: 421 ggatgaaaggaaatggagggctggtaagatgcaacaaggtgctcctagtgctgtggctcc 480 Query: 945 tacggttattg 955 ||||||||||| Sbjct: 481 tacggttattg 491>RGT4569_5.1 Length = 222 Score = 44.1 bits (22), Expect(2) = 1e-05 Identities = 22/22 (100%) Strand = Plus / Minus Query: 638 tgctgcagtgcctccttgtgct 659 |||||||||||||||||||||| Sbjct: 215 tgctgcagtgcctccttgtgct 194Score = 28.2 bits (14), Expect(2) = 1e-05 Identities = 23/26 (88%) Strand = Plus / Minus Query: 767 gcctgctggacccggcgatgctcttg 792 ||||||||||||| | || ||||||| Sbjct: 86 gcctgctggacccagagaagctcttg 61>RGT4568_5.1 Length = 222 Score = 44.1 bits (22), Expect(2) = 1e-05 Identities = 22/22 (100%) Strand = Plus / Minus Query: 638 tgctgcagtgcctccttgtgct 659 |||||||||||||||||||||| Sbjct: 215 tgctgcagtgcctccttgtgct 194Score = 28.2 bits (14), Expect(2) = 1e-05 Identities = 23/26 (88%) Strand = Plus / Minus Query: 767 gcctgctggacccggcgatgctcttg 792 ||||||||||||| | || ||||||| Sbjct: 86 gcctgctggacccagagaagctcttg 61>RdSpm2990B_3.1 Length = 915 Score = 48.1 bits (24), Expect = 4e-05 Identities = 24/24 (100%) Strand = Plus / Plus Query: 123 ggcggcggcggcggcggcggaggc 146 |||||||||||||||||||||||| Sbjct: 745 ggcggcggcggcggcggcggaggc 768>RGT3379_5.1 Length = 766 Score = 46.1 bits (23), Expect = 1e-04 Identities = 26/27 (96%) Strand = Plus / Minus Query: 123 ggcggcggcggcggcggcggaggccgg 149 |||||||||||||||||||| |||||| Sbjct: 312 ggcggcggcggcggcggcggcggccgg 286Database: combined_genbank_submitted Posted date: Mar 12, 2007 10:55 AM Number of letters in database: 4,449,403 Number of sequences in database: 10,808 Lambda K H 1.37 0.711 1.31 Matrix: blastn matrix:1 -3 Number of Hits to DB: 3649 Number of Sequences: 10808 Number of extensions: 3649 Number of successful extensions: 1347 Number of sequences better than 1.0e-04: 10 length of query: 1296 length of database: 4,449,403 effective HSP length: 16 effective length of query: 1280 effective length of database: 4,276,475 effective search space: 5473888000 effective search space used: 5473888000 T: 0 A: 0 X1: 6 (11.9 bits) X2: 15 (29.7 bits) S1: 12 (24.3 bits) S2: 23 (46.1 bits)